Virtual Cell Challenge 2025
Published:
The original challenge is annouced in cell as an commentary Virtual Cell Challenge: Toward a Turing test for the virtual cell
Abstract
Virtual cells are an emerging frontier at the intersection of artificial intelligence and biology. A key goal of these cell state models is predicting cellular responses to perturbations. The Virtual Cell Challenge is being established to catalyze progress toward this goal. This recurring and open benchmark competition from the Arc Institute will provide an evaluation framework, purpose-built datasets, and a venue for accelerating model development.
Introduction
Rapid advances in single-cell technologies and high-throughput perturbational measurements have motivated renewed efforts to model cell behavior quantitatively. These “virtual cells” are expected to learn the relationship between cell state and function and are intended to predict the consequences of perturbations—such as a gene knockdown or the application of a drug—across cell types and cell contexts. Given the central role of cellular behavior across diverse fields of basic biology and translational research, the impact of such models could be enormous. Initial efforts in the late 1990s and early 2000s aimed to create comprehensive computational models of the cell from first principles but stalled due to limited data availability, oversimplified assumptions, and computational constraints. Today, with massively improved single-cell measurement technologies, richer perturbational datasets, and advances in machine learning, the field is poised to revisit and realize these early ambitions. However, the absence of standardized benchmarks and evaluations for assessing model quality has limited progress in predictive cell state modeling. Motivated by the success of the CASP (Critical Assessment of protein Structure Prediction, https://predictioncenter.org) competition for protein folding, a structured, recurring community-wide competition can catalyze progress and align efforts around common goals. It is important to recognize that predictive models of cell behavior will differ from protein structure prediction in many ways. These models must account for additional complexity—such as the cell type, genetic background, and context of a cell—as well as the cellular phenotype being measured and predicted. This community challenge will serve to benchmark progress using carefully designed evaluation datasets and formalize standards with the goal of influencing the evolution of the field.
Open-source competitions can lead to rapid progress
Despite recent growth in the development of predictive models for single-cell perturbation responses, the research community working to model cellular states and behavior currently lacks principled shared evaluation frameworks that reflect the capacity of the model for biologically meaningful generalization across perturbations and cell types. Perturbation model performance is affected by substantial technical sources of variability in many existing datasets, including experimental noise introduced during the coupling of genetic perturbations with single-cell transcriptomic profiling, as well as the limited reproducibility of perturbation effects across independent experiments. Without standardized benchmarks and purpose-built evaluation datasets that evolve in real time alongside developments in the field, it is difficult to evaluate whether models are capturing generalizable biological structure rather than dataset-specific patterns. Past public competitions have laid important groundwork for benchmarking the prediction of cellular responses to perturbations, but key gaps remain. The Broad Institute’s Cancer Immunotherapy Data Science Grand Challenge, for instance, focused on predicting broad phenotypic shifts in T cells but did not assess responses at the resolution of gene expression. More recently, the 2023 NeurIPS-Kaggle competition advanced the field by evaluating gene expression changes in response to small-molecule perturbations in immune cells. Building on these efforts, the field is now ready for a benchmarking competition that assesses gene expression response to genetic perturbations—a task that is central to biological understanding of cell function. Unlike small molecules, genetic perturbations have precisely defined targets, making them ideal for probing causal gene-function relationships, even though the resulting transcriptional changes are often more subtle and harder to predict. To bridge these gaps, we are launching the Virtual Cell Challenge (https://virtualcellchallenge.org), an annual challenge that evaluates AI models of cellular response (Figure 1A). For the inaugural challenge, we have generated a dedicated dataset measuring single-cell responses to perturbations in a human embryonic stem cell line (H1 hESC). This set of perturbations was carefully curated to span a broad range of phenotypic responses, and experimental parameters were optimized to maximize the reproducibility of observed effects. The H1 hESC dataset generated for the Virtual Cell Challenge also contributes to the broader effort to establish experimental and quality control standards for reproducible, high-quality single-cell functional genomics (scFG) data. Such standards will enable progress and set the community up for building on a solid foundation.
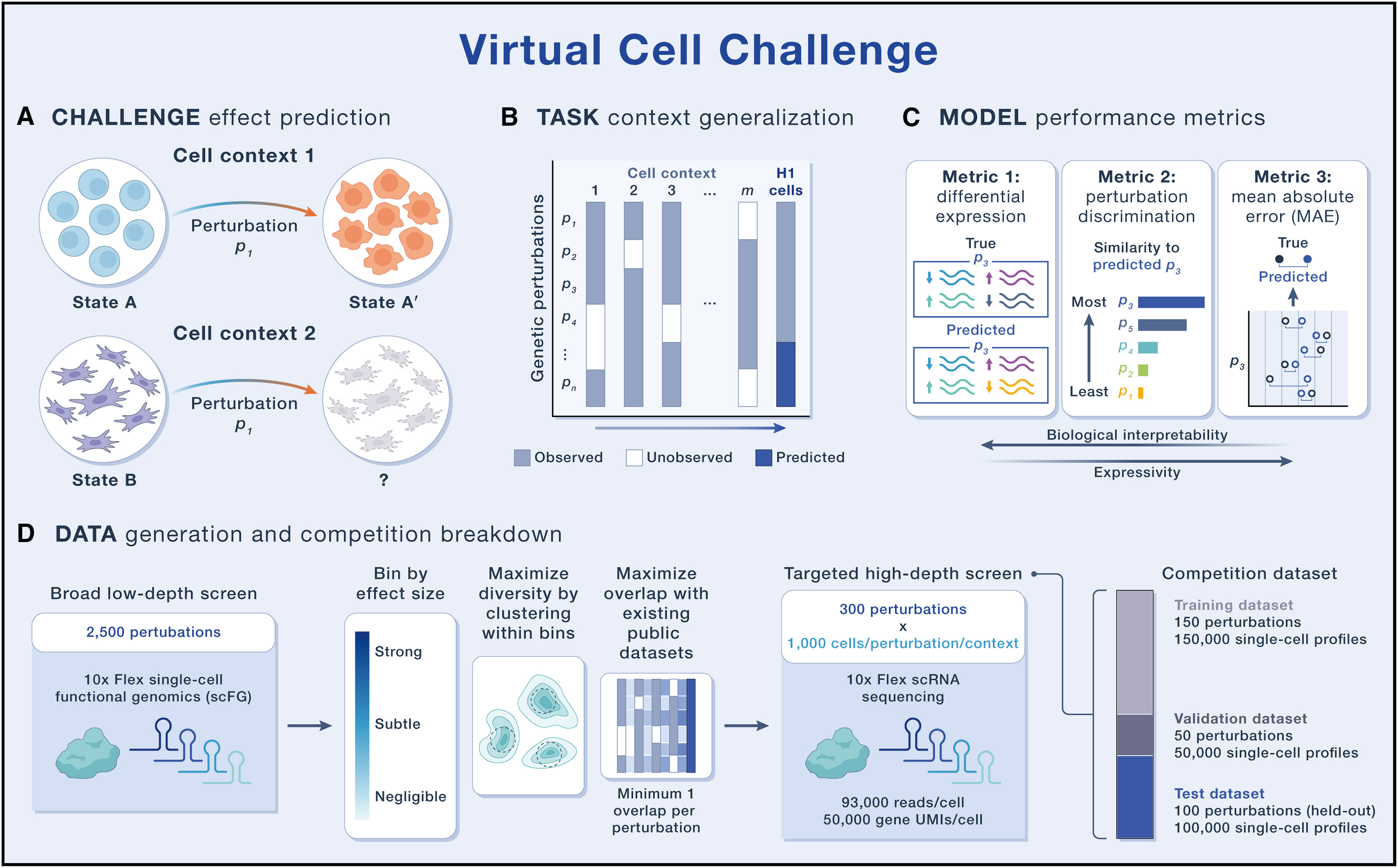 Figure 1 Overview of the Virtual Cell Challenge
Figure 1 Overview of the Virtual Cell Challenge
This effort is intended to create a level playing field, drive community engagement, and accelerate progress by providing high-quality benchmark datasets, a public leaderboard, and a mechanism for reproducible and fair comparison.
Format of the Virtual Cell Challenge
Task
Predictive models can be trained to generalize along several axes. Two core dimensions are (1) generalization across biological context (e.g., cell type, cell line, culture conditions, or even in vivo versus in vitro settings) and (2) generalization to novel genetic and/or chemical perturbations, including their combinations. The first Virtual Cell Challenge, launching in 2025, will focus on context generalization as a highly challenging real-world task: participants will predict the effects of single-gene perturbations in a held-out cell type (the H1 hESC; Figure 1B). The transcriptomic consequences of these genetic perturbations have been previously reported in at least one other cellular context. This reflects a common experimental reality: testing all perturbations in every context is impractical due to cost, yet accurate context-specific predictions are crucial because responses depend on factors like cell type, state, differentiation stage, culture conditions, and genetic background. Given that most published single-cell genetic perturbation datasets span only a handful of cell lines, true zero-shot generalization to new cell states is likely premature. A more appropriate strategy at this stage is few-shot adaptation, where a subset of perturbations in the new cellular context is provided to guide model generalization. To support this, we provide expression profiles for a subset of perturbations measured directly in H1 hESCs, enabling participants to adapt their models before predicting responses to the remaining unseen perturbations in the same cell type.
Evaluations
Evaluation metrics should reflect the core purpose of a virtual cell: simulating cellular behavior via in silico experiments—specifically, predicting gene expression responses to genetic perturbations. The main readouts from such an experiment are post-perturbation expression counts and differentially expressed gene sets. Based on these criteria, we have designed three metrics to evaluate model performance (Figure 1C). The first two, differential expression score and perturbation discrimination score, are complementary. The differential expression score evaluates how accurately a model predicts differential gene expression, a key output of most scFG experiments and an essential input for downstream biological interpretation. The perturbation discrimination score measures a model’s ability to distinguish between perturbations by ranking predictions according to their similarity to the true perturbational effect, regardless of their effect size. These two metrics capture distinct and nonoverlapping aspects of performance. For instance, a naive model that consistently predicts the same set of commonly differentially expressed genes from the training data might achieve a reasonable differential expression score. However, its perturbation discrimination score would be random or at the lower bound. Conversely, a model that successfully distinguishes perturbation effects based on subtle variation in an embedding space might perform well on perturbation discrimination. However, this is unlikely to produce biologically meaningful differentially expressed gene sets, which limits its practical utility as a replacement for experimental measurements. To ensure that predictions are also evaluated across all genes, including those that are not differentially expressed, we include a third metric: mean absolute error (MAE). While MAE is less biologically interpretable, it captures overall predictive accuracy and provides a global view of model performance across the entire gene expression profile. Together, these three metrics provide a comprehensive evaluation framework. We will use a combined score that appropriately weighs each component, and we will enforce minimum thresholds on all metrics to promote a balanced performance, discouraging models that perform well on one metric at the expense of the others. We will also report model ranking on each individual metric to aid with interpretation of results and highlight models that excel along any one dimension.
Datasets
The first Virtual Cell Challenge will be evaluated on Arc Institute-generated data for H1 hESCs. Additional training data are available from our Virtual Cell Atlas (https://arcinstitute.org/tools/virtualcellatlas) and public perturbation datasets.
New Arc-generated benchmark dataset
For this challenge, we used scFG to generate approximately 300,000 single-cell RNA-sequencing (scRNA-seq) profiles by silencing 300 carefully selected genes using CRISPR interference (CRISPRi) (Figure 1D). We selected these perturbations to represent a broad range in strength of downstream changes (number of significant differentially expressed genes) and phenotypic diversity of perturbation effects in our initial broad, low-depth screen in H1 hESCs while ensuring adequate representation in existing public datasets. The evaluation dataset was generated using the Flex scRNA-seq platform with high target cell coverage (median of ∼1,000 cells per perturbation) and depth of sequencing (over 50,000 unique molecular identifiers [UMIs] per cell). We adopted 10x Genomics’ Flex chemistry for this challenge because it offers a more scalable workflow, reduces batch effects through cell fixation, and achieves improved residual knockdown efficiency. Participants will receive a transcriptomic reference of unperturbed H1 hESCs, a training set consisting of single-cell profiles for 150 gene perturbations (∼150,000 cells), a validation set of 50 genes whose perturbations are used to create a live leaderboard during the competition, and a final test set of 100 held-out perturbations, released a week prior to the submission deadline. Final rankings will be based solely on performance on the held-out test set.
Arc Virtual Cell Atlas
This recently released resource is composed of scBaseCount and Tahoe-100M, the largest collection of publicly available observational and perturbational scRNA-seq datasets, respectively. With single-cell data from over 350 million cells and counting, this repository offers the community an ever-expanding training set for the next generation of virtual cell models.
Public perturbation datasets
We will also provide preprocessed versions of selected publicly available perturbation datasets that overlap with the perturbations that are included in the test set.11,16,17,18 While this selection is not exhaustive, these few large datasets with substantial perturbation overlaps are intended to support model training, since observing the effects of target perturbations in alternate cell contexts may be beneficial during model training.
Community engagement
The Virtual Cell Challenge was created to bring together researchers around the unified goal of building better models of cell behavior. Participants will receive regular updates via Arc’s Virtual Cell Challenge portal and be featured in leaderboard snapshots throughout the competition. By combining community engagement with rigorous benchmarking, our hope is that the competition will (1) surface best practices and critical data considerations—from training set quality to sequencing depth and platform choice—that will support more reproducible and robust research efforts; (2) identify methodological bottlenecks, clarify data and/or model choices, and converge on more reproducible standards for perturbation modeling; and (3) highlight the importance of training data quality, cell coverage, sequencing depth, and technology choice. By underscoring these issues, guidelines for more reproducible and principled data generation efforts might emerge.
Future directions
This effort is grounded in ongoing data generation at the Arc Institute, aimed at producing reference-quality datasets to advance the empirical foundations of predictive cell modeling. We hope it also encourages similar initiatives across the community to generate high-quality reference datasets, whether from scRNA-seq or other modalities, that contribute to building more comprehensive and generalizable models of cellular behavior. While the inaugural challenge focuses on genetic perturbation prediction in a single cell type, future challenges will expand to combinatorial perturbations and cross-cell-type generalization. Looking ahead, a comprehensive modeling of cellular behavior will require integrating diverse biological modalities—including transcriptional, proteomic, and epigenetic—across temporal and spatial dimensions in multicellular systems. Similarly, the inverse of perturbation response prediction, which aims to identify optimal perturbations that achieve a desired effect, is an important capability for Virtual Cell models with clear therapeutic relevance for future challenges. We view this initiative as an evolving test bed for advancing quantitative modeling, where both datasets and evaluation frameworks are designed to adapt as the field deepens its understanding of biological complexity. Metrics will be refined based on insights from prior years’ results. We invite the broader scientific community to shape and strengthen future versions of the Virtual Cell Challenge.
Conclusion
Virtual cells are poised to become foundational tools for biology, and to ensure that they reach their potential, we need clear and rigorous evaluations. The Virtual Cell Challenge aims to provide just that: a fair, open challenge to surface the best models, clarify the state of the art, and engage the community. We invite the community to engage with this first iteration and help better define the contours of predictive cellular modeling as a scientific discipline.
FAQ from the official website
What is the Virtual Cell Challenge?
The Virtual Cell Challenge is an annual competition hosted by Arc Institute to encourage advances in predictive cell modeling.
What dates do I need to know?
The 2025 Challenge runs from June to November. Winners will be announced in early December. Future years may have different timing. The dataset against which entries will be scored will be released on October 27, 2025 and you must submit a final entry by November 3, 2025.
Why this challenge and why now?
Understanding, predicting, and ultimately programming cellular responses to internal cues and external stimuli is a fundamental challenge in biology. Advances in single-cell technologies now enable large-scale measurements of cellular responses at the RNA level to genetic and chemical perturbations, fueling this exciting era of increasingly high quality biological data and predictive cellular modeling. The Virtual Cell Challenge seeks to create momentum for building and evaluating virtual cell models in the scientific community, for creating reference datasets, and attracting new contributors, especially in machine learning fields, to make progress in this area.
Where can I find more background on this effort?
Read more about the Virtual Cell Challenge in our commentary in Cell and about Arc’s first virtual cell model in this preprint.
Can I share my progress on social media?
Feel free to share about your experience.
I’m a researcher or student. Can I publish my results?
We encourage publishing and sharing research with the scientific community, if that is allowed by your employer or academic institution.
What will happen in future years?
The Virtual Cell Challenge will be an annual competition. We expect to release more dedicated data in future years that will help improve cell state models. We hope others in the field will also make progress in creating more and higher quality datasets, and in defining the most meaningful metrics for model performance.
Participation
Who can participate in the Challenge?
Teams from one to eight individuals may participate. You may participate in your personal capacity, or as an employee of your organization. Full details, including eligibility exceptions, are in the rules here.
Can I enter multiple models in the Challenge?
Individuals can only be part of one team, and each team may only submit one result per day. If a team would like to try multiple models, you may do so by generating results from different models on different days, but you must ultimately choose one model to generate results for your final entry that will be considered for prizes.
Can my employer or lab field multiple teams?
Yes. One lab or institution may field multiple teams with members who do not overlap.
How do I get permission from my employer to enter the Challenge? What if I am a student?
If you want to enter on behalf of your employer, you must take whatever steps your employer requires to get this permission. The process will vary depending on your organization. By entering the Challenge on behalf of your employer, you are confirming to us that you got that permission. We may contact your employer to double check. If you are a student at a college or university, the same approach applies: follow the permission processes at your organization if you want to enter on the organization’s behalf. A good place to start is with your faculty advisor.
Legal
What are the rules of the Challenge?
You can review the rules here.
Can I use the H1 hESC Challenge datasets that Arc has created for other purposes outside of the Challenge?
Yes. The datasets will be openly licensed via CC0 1.0, as is the case with the Tahoe-100M and scBaseCount datasets on the Arc Virtual Cell Atlas.
Can I use proprietary models or data to generate results for the Challenge?
Yes, so long as you have the right to do so.
Am I required to submit my code to the Challenge?
Participants will run their own models to generate results, and only those results form your entry. We will not require model code to be submitted for verification in all cases, but winning submissions are subject to competition rules and we reserve the right to eliminate suspicious submissions and/or request more information for verification. If you are a finalist, to be eligible to win a prize, you must submit a description we will share with the public about how your model works, at least at a high level, including some discussion of the model architecture and training approach. This description must include how, if at all, you used Arc’s State Model in your participation in the Challenge.
Will Arc own my model?
No. Arc does not obtain any rights to your model by virtue of your participation in the Challenge.
Can I use Arc’s State model in the Challenge?
If you are a noncommercial entrant, yes, you may use Arc’s State Model in order to participate in the Challenge (and such use will be considered a Non-Commercial Purpose solely to the extent of your use for participation in the Challenge), provided that you comply in all respects with all terms, conditions or restrictions in the Arc Research Institute State Model Non-Commercial License.
Teams
How many people can be on a team?
Each team may have up to 8 members, including a team lead.
Can I be part of multiple teams?
No. Each individual must choose one team to be part of the competition.
Can I participate as an individual separate from my employer?
The Challenge is open to both institutional teams and individuals. You should understand your own employment terms in order to determine if you can participate as an individual.
Can individuals from multiple universities enter the Challenge as a team?
Yes. See the rules for more details.
How do I change teams?
Contact help@virtualcellchallenge.org to make this request.
What should I do if my team wants to join forces with another team?
Contact help@virtualcellchallenge.org to make this request.
Troubleshooting
What should I do if I have a problem with my submission or other issue with the website?
Please email help@virtualcellchallenge.org
How can I connect and discuss the competition with other participants?
Registered participants can join a Discord community we have created for the Challenge.
Data
What data sets are part of the Challenge?
Participants in the Challenge get access to three parts of a new, experimentally-generated perturbation data set. The training set can be used to fine tune a virtual cell model, the validation set is used to run inference against your model in order to generate predictions for the leaderboard, and the final test set will be released in October and will be used for final predictions upon which winners will be selected. Winners will be announced in December. Read more about the data here.
Can I use other data to train or fine tune my model?
As long as you have the appropriate permissions to use the data, you may use any data to improve the performance of your predictions. This includes proprietary data you have access to. The Challenge will not need access to your data for the competition.
Scoring
How are scores calculated in the code provided by the Challenge?
The code for calculating the scores is contained in the cell-eval package. The README file (Run and Score sections) gives the exact commands that we run to calculate the metrics. The pdex package calculates p-values and log-fold-changes using the same method as scanpy rank_genes_group function with the Wilcoxon option.
Prizes
I’m outside the United States, am I eligible to win a prize?
You are eligible to win a prize if you are eligible to participate in the Challenge, under the rules here.
What happens if I am a prize winner?
We will be in contact with finalists in advance of prize announcements to share further details, such as confirming eligibility, information needed for tax reporting, and announcement plans. See also the rules for more information.
Do I have to pay taxes on my prize earnings?
Consult with a tax expert to help you understand the implications of the prizes.
How are prizes divided among team members?
The prize for a winning team will be paid in equal shares to all team members identified in the finalist confirmations.
